Screening Assistant 2 v1.1
HTS flags
Last edited: 02/11/2011
By: VLG
Sourceforge: SA2 website
Help: SA2 forums
Documentation index
This page describes the HTS flags available in SA2.
Table of Contents
Overview - top
SA2 was primarilly designed to manage screening libraries. In the past two decades, screening assays has progressively evolved from cell-based biological assays to high-throughput biochemical assay platform which enable to test much more compounds in a single run. Although these new technologies alows a much higher throughput, they also have their own caveats and artifacts that have been extensively described in the litterature [1-3]. For example, Rishton [1, 2] provides various example of such artifacts, i.e. nucleophilic protease assays that have been shown particularly sensitive to protein-reactive electrophiles, or phosphatase assays are particularly sensitive to phosphorylated compounds, salts, metals and polyions. Baell & Holloway [3] recently described a set of classes of molecules that are likely to be turned into false positives in biochemical screenings. They advise to remove such compounds from screening libraries to avoid waste of time and ressources.
SA2 makes it possible to compute a set of flags intended to warn the user on such potentially problematic compounds. Let's describe them a bit.
Reactive
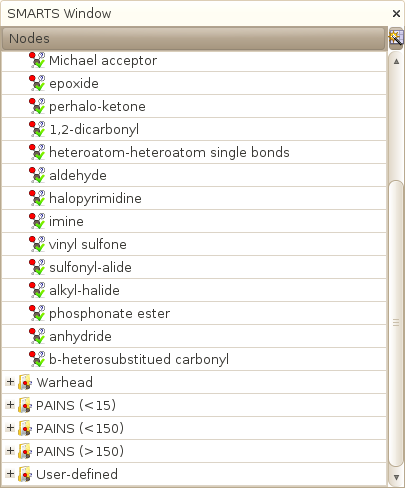
Reactives are those compounds containing electrophilic functional groups that are known to be protein-reactive covalent-acting molecules, and which usually lead to false positive in biochemical screening. There are 20 reactive patterns in SA2 which have been extracted from [1]. A compound that matches any of the 20 patterns will be flagged as reactive.
Warhead
Warheads are another class of compounds which are known to be reversible and non-covalent binders which also exhibit false positive behaviors because of there reactivity. There are 14 warhead patterns in SA2, which have been extracted from [1] too.
Pan Assay Interference Compounds (PAINS)
Three classes of compounds have been defined by Baell and Holloway [3] as being frequently found as false positive in biochemical assays. The three classes represent :
- PAINSl15: these are classes of compounds that encode for less than 15 compounds in one or more family in [3]. These patterns are therefore quite specific, and actually numerous (more than 400).
- PAINSl150: these are classes of compounds that encode between 15 and 150 compounds in one or more family in [3].
- PAINSl15: classes of compounds that encoded for more than 150 compounds in one or more family in [3].
Druglikeness
The famous Lipinski RO5 flag will also be computed [4]. No need to further explain this I guess :).
Fragment-likeness
A fragment likness flag (RO3) will be computed too. The rules used are those of Congreve & al [5], with no violation allowed (as they do not specify it in their mini-article).
Customising HTS flags - top
Because artifacts are often protein or assay dependant, SA2 offers the possibility to edit the existing SMARTS patterns, by desactivating those that you find inapropriate for your problem, or by deleting them directely (irreversible!). You can also add your own SMARTS for each flag, and in particular, create your own set of smarts that will be matched to compute the User-defined HTS flag (that by default do not contain any SMARTS pattern). These patterns can of course represent anything, from your own PAINS SMARTS, to toxicophores or such...
To do so, you will have to use the SMARTS List window.
Note that if you perform any modification on the SMARTS, you will have to recalcualte the values of each flag in the database so the flags values remain consistent with your modifications. You can do so using the Database->Admin->Recalculate Flags menu.
References - top
[1] Rishton, G.M.: Reactive compounds and in vitro false positives. in HTS. Drug Discov. Today 1997, 2:382–384
[2] Rishton, G.M.: Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov. Today 2003, 8(2):86–96.
[3] Baell JB, Holloway GA: New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of Medicinal Chemistry 2010, 53(7):2719–2740.
[4] Lipinski CA, Lombardo F, Dominy BW, Feeney PJ: Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews 2001, 46(1-3):3–26.
[5] Congreve M, Carr R, Murray C, Jhoti H: A ’rule of three’ for fragment-based lead discovery? Drug Discovery Today 2003, 8(19):876–877.